In order to cure disease, we need to remember we are also animals
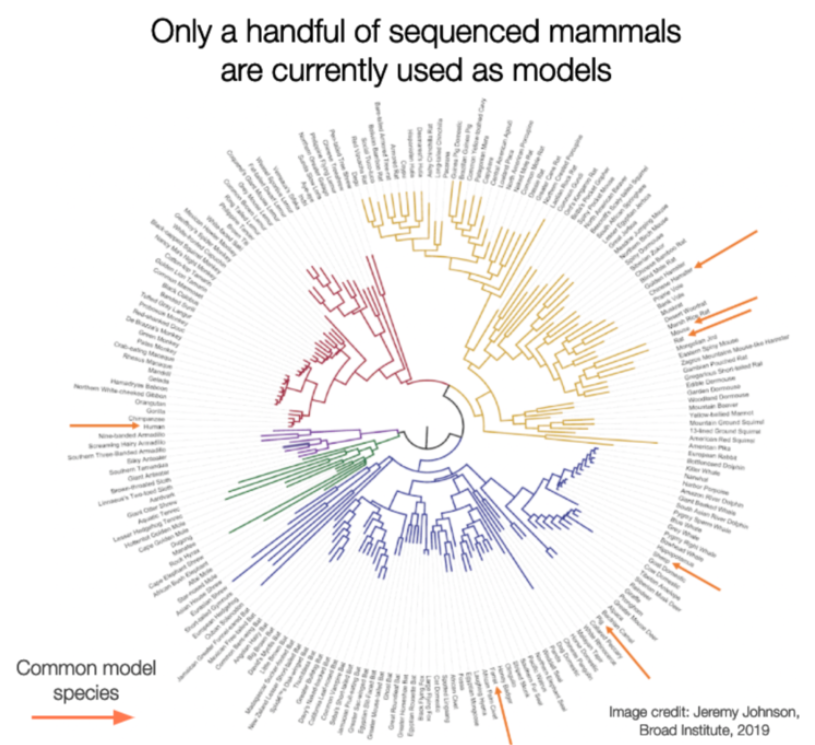
For the past several decades biomedical drug discovery has been a bit like the joke about a drunk man looking for his keys under the lamppost. This is because, until recently, (within the last 5-10 years or so), we understood the genomes of only 4-5 model species. Therefore, we used genetic modifications to tweak these species to be more like the “human” diseases we wanted to study and then, were disappointed when insights from these wrangled models didn’t translate into human therapies. These failures cost billions of dollars and limited the new therapies developed for patients with a range of diseases including heart disease, diabetes, Alzheimer’s, and others. With the precipitous drop in sequencing costs for DNA and RNA and the availability of new, high quality animal genomes for only a few thousand dollars, there are literally hundreds of species now available to us to explore biological processes and help us understand mechanisms of disease, and resistance to disease, that are core to our biology.
This makes for interesting dinner conversation, but more importantly, it highlights a critical failing underpinning our biomedical research complex. Recent research demonstrates that evolutionary conservation in mammals is the best predictor of function, and highly conserved genetic targets advance further in clinical development than targets that have evolved more recently.[source][source][source]
Even the greenest of biology students quickly learns to turn on those conservation tracks on the UCSC browser in order to check the conservation of their favorite gene from their latest sequencing studies and to prioritize what is important from gene lists. It’s intuitive that genes and pathways conserved over hundreds of millions of years of evolution are probably doing important things.
However, what is missing from this approach is an understanding of how those genes, and subsequently proteins, are changing in animals in conditions that mimic or better model human diseases than the mice, rats, fish, flies and worms we use today. In short, functional genomics.
And this is not at all to suggest that research in mice, rats, fish, flies and worms is not important! To the contrary, much of our understanding of the function of single genes is due to studies in these species where we disable single genes and evaluate the resulting effects on model species. However, there are only 8,000 or single gene (Mendelian) disorders that affect humans and many of these disorders affect perhaps only a few hundred or thousand people in the world.
Most diseases are complex and involve interactions of multiple genes in multiple tissues reacting to a set of external environmental cues or risk factors. Attempting to induce a similar set of conditions in an ill-suited animal model is pretty well doomed to fail in terms of predicting treatment responses in people.
Animals, like 13-lined ground squirrels that survive the human equivalent of 25 heart attacks a year and don't end up needing a heart transplant, animals that live 10x longer than their peers, but almost never get cancer, animals that dive to the deepest depths of the ocean and are protected from severe lack of oxygen, animals whose body temperatures dip below freezing, live to tell the tale and can teach us about organ and tissue preservation in extreme settings.
There are rockstar researchers out there who have spent their entire careers understanding these amazing species, building research colonies, creeping into bear caves in the dead of winter, and chasing seals across the Arctic circle (we know, one of these researchers helped start this company and others are on our advisory board!)
These researchers are now taking advantage of the genomic revolution to use high-throughput sequencing technologies to understand the genes involved in the amazing traits they observe. This data is out there, this data is extremely valuable to help us understand how our genes function in health and disease.
However, this data is scattered and unorganized, in genomics repositories, under-utilized tissue banks, in supplemental tables, on hard drives. In order to realize the potential of this emerging resource, we need to bring it together, examine the consistent trends across organisms, and align that data to existing genomic resources in human and other model organism data. Enter Fauna Bio.
My mother always told me everything happens for a reason. For the three co-founders of Fauna, it was a lot of unexpected twists and turns that led us to end up together at Stanford in the fall of 2017. We come from different corners of biology; veterinary science (Ashley), comparative genomics(Linda), and hibernation biology(Katie). And we combine these skillsets for a unique sense of what is possible with the data revolution beginning now in “non-model” organisms.
We are honored to have raised 4.1M in seed funding from strong and aligned partners to advance our goals. If we succeed in our mission to bring non-model organisms into the mainstream of biological discovery, the idea of a “model organism” will be a construct of history and of a time when we only had the capacity to understand the biology of species where we controlled their genes. Now we know all animals can be models, and nearly every human disease can be modeled in one or more species. It’s a matter of finding the right model. This false separation of “human” and “animal” data is an antiquated notion. It’s time to remember that humans ARE animals and by ignoring many of our relatives, we cannot truly understand ourselves.